Computational Protein Design Overview
Published:
Computational Protein Design(CPD) has produced impressive results for engineering new proteins, resulting in a wide variety of applications. In this blog, the whole pipeline and mainstream methods of CPD will be concluded.
Problem Statement
Aim
The aim of CPD is to design proteins with new or enhanced properties, which may include thermostability, binding affinity, ligand specificity and new activities. We should not only predict a sequence folding onto a target structure, but also bestow the structure a specific function and required properties.
Formulation
The most usual approach to CPD consists in choosing or de novo constructing a target backbone structure that could carry the function of interest and then identify a sequence that will:
- fold onto this backbone
- present the target properties
In this case, this black-box could be defined as a calculator with:
- input: target backbone with target properties
- output: designed sequence
Actually, the CPD problem can be formulated as an optimization problem: given a input backbone, find a sequence that:
- maximally stabilizes the input backbone
- fulfill the desired function
by minimizing a score function that usually combines the free energy of the resulting protein with other function-related criteria.
Difficulty
Protein data representation and neural architecture selection
As for DL methods, the chosen input representation must be adapted to protein of variable lengths and be able to concisely encode the relational information of the protein structure.
Since protein structures are naturally insensitive to translations and rotations, only the relative orientations and position of structural motifs matters locally. Ideally, the protein structure representation should account for these properties to make training more efficient. However, for now, there is no final consensus about which representation is best to capture protein structures and which neural architecture should be privileged.
CPD is an ill-posed problem
Sometimes the sequence is optimized for the target structure, but this structure may not be optimal for the sequence which ultimately may fold in a different structure.
In this case, following the design itself, it is usual to filter designed sequences to keep only those that are reliably predicted to fold in the target structure, a procedure called forward folding.
Pipeline
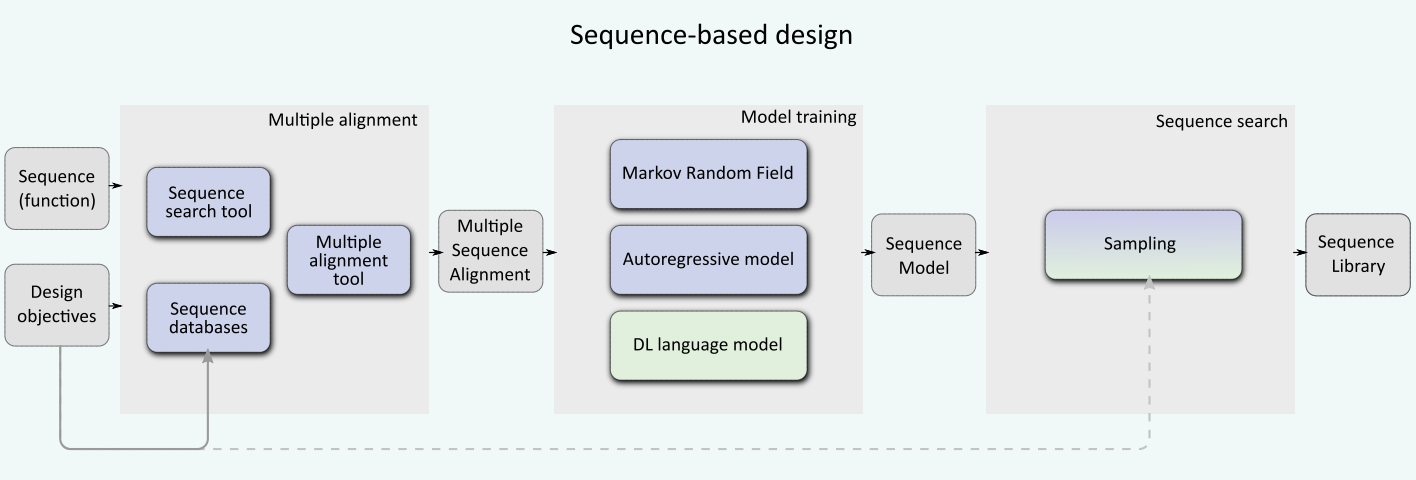
Sequence-based
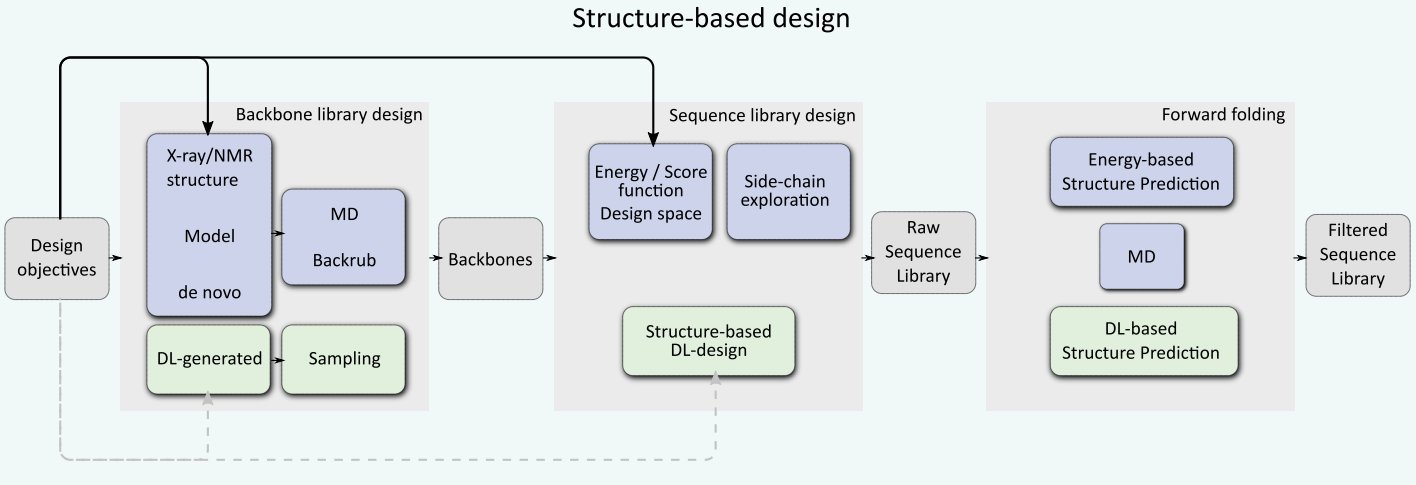
Structure-based
Reference
https://www.biorxiv.org/content/10.1101/2022.08.31.505981v1.full.pdf
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8584038/pdf/ijms-22-11741.pdf
